8. Elementary Probability with Matrices#
This lecture uses matrix algebra to illustrate some basic ideas about probability theory.
After introducing underlying objects, we’ll use matrices and vectors to describe probability distributions.
Among concepts that we’ll be studying include
a joint probability distribution
marginal distributions associated with a given joint distribution
conditional probability distributions
statistical independence of two random variables
joint distributions associated with a prescribed set of marginal distributions
couplings
copulas
the probability distribution of a sum of two independent random variables
convolution of marginal distributions
parameters that define a probability distribution
sufficient statistics as data summaries
We’ll use a matrix to represent a bivariate or multivariate probability distribution and a vector to represent a univariate probability distribution
This companion lecture describes some popular probability distributions and describes how to use Python to sample from them.
In addition to what’s in Anaconda, this lecture will need the following libraries:
!pip install prettytable
As usual, we’ll start with some imports
import numpy as np
import matplotlib.pyplot as plt
import prettytable as pt
from mpl_toolkits.mplot3d import Axes3D
from matplotlib_inline.backend_inline import set_matplotlib_formats
set_matplotlib_formats('retina')
8.1. Sketch of Basic Concepts#
We’ll briefly define what we mean by a probability space, a probability measure, and a random variable.
For most of this lecture, we sweep these objects into the background
Note
Nevertheless, they’ll be lurking beneath induced distributions of random variables that we’ll focus on here. These deeper objects are essential for defining and analysing the concepts of stationarity and ergodicity that underly laws of large numbers. For a relatively nontechnical presentation of some of these results see this chapter from Lars Peter Hansen and Thomas J. Sargent’s online monograph titled “Risk, Uncertainty, and Values”:https://lphansen.github.io/QuantMFR/book/1_stochastic_processes.html.
Let
Let
Let
The pair
A probability measure
this is the “probability” that
A random variable
The random variable
where
We call this the induced probability distribution of random variable
Instead of working explicitly with an underlying probability space
That is how we’ll proceed in this lecture and in many subsequent lectures.
8.2. What Does Probability Mean?#
Before diving in, we’ll say a few words about what probability theory means and how it connects to statistics.
We also touch on these topics in the quantecon lectures https://python.quantecon.org/prob_meaning.html and https://python.quantecon.org/navy_captain.html.
For much of this lecture we’ll be discussing fixed “population” probabilities.
These are purely mathematical objects.
To appreciate how statisticians connect probabilities to data, the key is to understand the following concepts:
A single draw from a probability distribution
Repeated independently and identically distributed (i.i.d.) draws of “samples” or “realizations” from the same probability distribution
A statistic defined as a function of a sequence of samples
An empirical distribution or histogram (a binned empirical distribution) that records observed relative frequencies
The idea that a population probability distribution is what we anticipate relative frequencies will be in a long sequence of i.i.d. draws. Here the following mathematical machinery makes precise what is meant by anticipated relative frequencies
Law of Large Numbers (LLN)
Central Limit Theorem (CLT)
Scalar example
Let
where
We sometimes write
as a short-hand way of saying that the random variable
Consider drawing a sample
What do the “identical” and “independent” mean in IID or iid (“identically and independently distributed”)?
“identical” means that each draw is from the same distribution.
“independent” means that joint distribution equal products of marginal distributions, i.e.,
We define an empirical distribution as follows.
For each
Key concepts that connect probability theory with statistics are laws of large numbers and central limit theorems
LLN:
A Law of Large Numbers (LLN) states that
CLT:
A Central Limit Theorem (CLT) describes a rate at which
Remarks
For “frequentist” statisticians, anticipated relative frequency is all that a probability distribution means.
But for a Bayesian it means something else – something partly subjective and purely personal.
we say “partly” because a Bayesian also pays attention to relative frequencies
8.3. Representing Probability Distributions#
A probability distribution
Sometimes, but not always, a random variable can also be described by density function
Here
When a probability density exists, a probability distribution can be characterized either by its CDF or by its density.
For a discrete-valued random variable
the number of possible values of
we replace a density with a probability mass function, a non-negative sequence that sums to one
we replace integration with summation in the formula like (8.1) that relates a CDF to a probability mass function
In this lecture, we mostly discuss discrete random variables.
Doing this enables us to confine our tool set basically to linear algebra.
Later we’ll briefly discuss how to approximate a continuous random variable with a discrete random variable.
8.4. Univariate Probability Distributions#
We’ll devote most of this lecture to discrete-valued random variables, but we’ll say a few things about continuous-valued random variables.
8.4.1. Discrete random variable#
Let
Here, we choose the maximum index
Define
for which
This vector defines a probability mass function.
The distribution (8.2)
has parameters
These parameters pin down the shape of the distribution.
(Sometimes
Such a “non-parametric” distribution has as many “parameters” as there are possible values of the random variable.
We often work with special distributions that are characterized by a small number parameters.
In these special parametric distributions,
where
Remarks:
A statistical model is a joint probability distribution characterized by a list of parameters
The concept of parameter is intimately related to the notion of sufficient statistic.
A statistic is a nonlinear function of a data set.
Sufficient statistics summarize all information that a data set contains about parameters of statistical model.
Note that a sufficient statistic corresponds to a particular statistical model.
Sufficient statistics are key tools that AI uses to summarize or compress a big data set.
R. A. Fisher provided a rigorous definition of information – see https://en.wikipedia.org/wiki/Fisher_information
An example of a parametric probability distribution is a geometric distribution.
It is described by
Evidently,
Let
8.4.2. Continuous random variable#
Let
where
8.5. Bivariate Probability Distributions#
We’ll now discuss a bivariate joint distribution.
To begin, we restrict ourselves to two discrete random variables.
Let
Then their joint distribution is described by a matrix
whose elements are
where
8.6. Marginal Probability Distributions#
The joint distribution induce marginal distributions
For example, let a joint distribution over
The implied marginal distributions are:
Digression: If two random variables
8.7. Conditional Probability Distributions#
Conditional probabilities are defined according to
where
For a pair of discrete random variables, we have the conditional distribution
where
Note that
Remark: The mathematics of conditional probability implies:
Note
Formula (8.4) is also what a Bayesian calls Bayes’ Law. A Bayesian statistician regards marginal probability distribution
For the joint distribution (8.3)
8.8. Transition Probability Matrix#
Consider the following joint probability distribution of two random variables.
Let
where
An associated conditional distribution is
We can define a transition probability matrix
where
The first row is the probability that
The second row is the probability that
Note that
8.9. Application: Forecasting a Time Series#
Suppose that there are two time periods.
Let
Suppose that
A conditional distribution is
Remark:
This formula is a workhorse for applied economic forecasters.
8.10. Statistical Independence#
Random variables X and Y are statistically independent if
where
Conditional distributions are
8.11. Means and Variances#
The mean and variance of a discrete random variable
A continuous random variable having density
8.12. Matrix Representations of Some Bivariate Distributions#
Let’s use matrices to represent a joint distribution, conditional distribution, marginal distribution, and the mean and variance of a bivariate random variable.
The table below illustrates a probability distribution for a bivariate random variable.
Marginal distributions are
Sampling:
Let’s write some Python code that let’s us draw some long samples and compute relative frequencies.
The code will let us check whether the “sampling” distribution agrees with the “population” distribution - confirming that the population distribution correctly tells us the relative frequencies that we should expect in a large sample.
# specify parameters
xs = np.array([0, 1])
ys = np.array([10, 20])
f = np.array([[0.3, 0.2], [0.1, 0.4]])
f_cum = np.cumsum(f)
# draw random numbers
p = np.random.rand(1_000_000)
x = np.vstack([xs[1]*np.ones(p.shape), ys[1]*np.ones(p.shape)])
# map to the bivariate distribution
x[0, p < f_cum[2]] = xs[1]
x[1, p < f_cum[2]] = ys[0]
x[0, p < f_cum[1]] = xs[0]
x[1, p < f_cum[1]] = ys[1]
x[0, p < f_cum[0]] = xs[0]
x[1, p < f_cum[0]] = ys[0]
print(x)
[[ 1. 0. 1. ... 0. 0. 1.]
[20. 10. 20. ... 20. 10. 10.]]
Note
To generate random draws from the joint distribution
# marginal distribution
xp = np.sum(x[0, :] == xs[0])/1_000_000
yp = np.sum(x[1, :] == ys[0])/1_000_000
# print output
print("marginal distribution for x")
xmtb = pt.PrettyTable()
xmtb.field_names = ['x_value', 'x_prob']
xmtb.add_row([xs[0], xp])
xmtb.add_row([xs[1], 1-xp])
print(xmtb)
print("\nmarginal distribution for y")
ymtb = pt.PrettyTable()
ymtb.field_names = ['y_value', 'y_prob']
ymtb.add_row([ys[0], yp])
ymtb.add_row([ys[1], 1-yp])
print(ymtb)
marginal distribution for x
+---------+---------------------+
| x_value | x_prob |
+---------+---------------------+
| 0 | 0.500384 |
| 1 | 0.49961599999999995 |
+---------+---------------------+
marginal distribution for y
+---------+----------+
| y_value | y_prob |
+---------+----------+
| 10 | 0.400001 |
| 20 | 0.599999 |
+---------+----------+
# conditional distributions
xc1 = x[0, x[1, :] == ys[0]]
xc2 = x[0, x[1, :] == ys[1]]
yc1 = x[1, x[0, :] == xs[0]]
yc2 = x[1, x[0, :] == xs[1]]
xc1p = np.sum(xc1 == xs[0])/len(xc1)
xc2p = np.sum(xc2 == xs[0])/len(xc2)
yc1p = np.sum(yc1 == ys[0])/len(yc1)
yc2p = np.sum(yc2 == ys[0])/len(yc2)
# print output
print("conditional distribution for x")
xctb = pt.PrettyTable()
xctb.field_names = ['y_value', 'prob(x=0)', 'prob(x=1)']
xctb.add_row([ys[0], xc1p, 1-xc1p])
xctb.add_row([ys[1], xc2p, 1-xc2p])
print(xctb)
print("\nconditional distribution for y")
yctb = pt.PrettyTable()
yctb.field_names = ['x_value', 'prob(y=10)', 'prob(y=20)']
yctb.add_row([xs[0], yc1p, 1-yc1p])
yctb.add_row([xs[1], yc2p, 1-yc2p])
print(yctb)
conditional distribution for x
+---------+--------------------+--------------------+
| y_value | prob(x=0) | prob(x=1) |
+---------+--------------------+--------------------+
| 10 | 0.7501806245484386 | 0.2498193754515614 |
| 20 | 0.3338522230870385 | 0.6661477769129616 |
+---------+--------------------+--------------------+
conditional distribution for y
+---------+---------------------+--------------------+
| x_value | prob(y=10) | prob(y=20) |
+---------+---------------------+--------------------+
| 0 | 0.5996854415808659 | 0.4003145584191341 |
| 1 | 0.20000960737846665 | 0.7999903926215334 |
+---------+---------------------+--------------------+
Let’s calculate population marginal and conditional probabilities using matrix algebra.
(1) Marginal distribution:
(2) Conditional distribution:
These population objects closely resemble the sample counterparts computed above.
Let’s wrap some of the functions we have used in a Python class that will let us generate and sample from a discrete bivariate joint distribution.
class discrete_bijoint:
def __init__(self, f, xs, ys):
'''initialization
-----------------
parameters:
f: the bivariate joint probability matrix
xs: values of x vector
ys: values of y vector
'''
self.f, self.xs, self.ys = f, xs, ys
def joint_tb(self):
'''print the joint distribution table'''
xs = self.xs
ys = self.ys
f = self.f
jtb = pt.PrettyTable()
jtb.field_names = ['x_value/y_value', *ys, 'marginal sum for x']
for i in range(len(xs)):
jtb.add_row([xs[i], *f[i, :], np.sum(f[i, :])])
jtb.add_row(['marginal_sum for y', *np.sum(f, 0), np.sum(f)])
print("\nThe joint probability distribution for x and y\n", jtb)
self.jtb = jtb
def draw(self, n):
'''draw random numbers
----------------------
parameters:
n: number of random numbers to draw
'''
xs = self.xs
ys = self.ys
f_cum = np.cumsum(self.f)
p = np.random.rand(n)
x = np.empty([2, p.shape[0]])
lf = len(f_cum)
lx = len(xs)-1
ly = len(ys)-1
for i in range(lf):
x[0, p < f_cum[lf-1-i]] = xs[lx]
x[1, p < f_cum[lf-1-i]] = ys[ly]
if ly == 0:
lx -= 1
ly = len(ys)-1
else:
ly -= 1
self.x = x
self.n = n
def marg_dist(self):
'''marginal distribution'''
x = self.x
xs = self.xs
ys = self.ys
n = self.n
xmp = [np.sum(x[0, :] == xs[i])/n for i in range(len(xs))]
ymp = [np.sum(x[1, :] == ys[i])/n for i in range(len(ys))]
# print output
xmtb = pt.PrettyTable()
ymtb = pt.PrettyTable()
xmtb.field_names = ['x_value', 'x_prob']
ymtb.field_names = ['y_value', 'y_prob']
for i in range(max(len(xs), len(ys))):
if i < len(xs):
xmtb.add_row([xs[i], xmp[i]])
if i < len(ys):
ymtb.add_row([ys[i], ymp[i]])
xmtb.add_row(['sum', np.sum(xmp)])
ymtb.add_row(['sum', np.sum(ymp)])
print("\nmarginal distribution for x\n", xmtb)
print("\nmarginal distribution for y\n", ymtb)
self.xmp = xmp
self.ymp = ymp
def cond_dist(self):
'''conditional distribution'''
x = self.x
xs = self.xs
ys = self.ys
n = self.n
xcp = np.empty([len(ys), len(xs)])
ycp = np.empty([len(xs), len(ys)])
for i in range(max(len(ys), len(xs))):
if i < len(ys):
xi = x[0, x[1, :] == ys[i]]
idx = xi.reshape(len(xi), 1) == xs.reshape(1, len(xs))
xcp[i, :] = np.sum(idx, 0)/len(xi)
if i < len(xs):
yi = x[1, x[0, :] == xs[i]]
idy = yi.reshape(len(yi), 1) == ys.reshape(1, len(ys))
ycp[i, :] = np.sum(idy, 0)/len(yi)
# print output
xctb = pt.PrettyTable()
yctb = pt.PrettyTable()
xctb.field_names = ['x_value', *xs, 'sum']
yctb.field_names = ['y_value', *ys, 'sum']
for i in range(max(len(xs), len(ys))):
if i < len(ys):
xctb.add_row([ys[i], *xcp[i], np.sum(xcp[i])])
if i < len(xs):
yctb.add_row([xs[i], *ycp[i], np.sum(ycp[i])])
print("\nconditional distribution for x\n", xctb)
print("\nconditional distribution for y\n", yctb)
self.xcp = xcp
self.xyp = ycp
Let’s apply our code to some examples.
Example 1
# joint
d = discrete_bijoint(f, xs, ys)
d.joint_tb()
The joint probability distribution for x and y
+--------------------+-----+--------------------+--------------------+
| x_value/y_value | 10 | 20 | marginal sum for x |
+--------------------+-----+--------------------+--------------------+
| 0 | 0.3 | 0.2 | 0.5 |
| 1 | 0.1 | 0.4 | 0.5 |
| marginal_sum for y | 0.4 | 0.6000000000000001 | 1.0 |
+--------------------+-----+--------------------+--------------------+
# sample marginal
d.draw(1_000_000)
d.marg_dist()
marginal distribution for x
+---------+----------+
| x_value | x_prob |
+---------+----------+
| 0 | 0.499565 |
| 1 | 0.500435 |
| sum | 1.0 |
+---------+----------+
marginal distribution for y
+---------+----------+
| y_value | y_prob |
+---------+----------+
| 10 | 0.399873 |
| 20 | 0.600127 |
| sum | 1.0 |
+---------+----------+
# sample conditional
d.cond_dist()
conditional distribution for x
+---------+--------------------+--------------------+-----+
| x_value | 0 | 1 | sum |
+---------+--------------------+--------------------+-----+
| 10 | 0.7498955918504125 | 0.2501044081495875 | 1.0 |
| 20 | 0.3327662311477404 | 0.6672337688522596 | 1.0 |
+---------+--------------------+--------------------+-----+
conditional distribution for y
+---------+---------------------+---------------------+-----+
| y_value | 10 | 20 | sum |
+---------+---------------------+---------------------+-----+
| 0 | 0.6002482159478747 | 0.39975178405212536 | 1.0 |
| 1 | 0.19984613386353872 | 0.8001538661364613 | 1.0 |
+---------+---------------------+---------------------+-----+
Example 2
xs_new = np.array([10, 20, 30])
ys_new = np.array([1, 2])
f_new = np.array([[0.2, 0.1], [0.1, 0.3], [0.15, 0.15]])
d_new = discrete_bijoint(f_new, xs_new, ys_new)
d_new.joint_tb()
The joint probability distribution for x and y
+--------------------+---------------------+------+---------------------+
| x_value/y_value | 1 | 2 | marginal sum for x |
+--------------------+---------------------+------+---------------------+
| 10 | 0.2 | 0.1 | 0.30000000000000004 |
| 20 | 0.1 | 0.3 | 0.4 |
| 30 | 0.15 | 0.15 | 0.3 |
| marginal_sum for y | 0.45000000000000007 | 0.55 | 1.0 |
+--------------------+---------------------+------+---------------------+
d_new.draw(1_000_000)
d_new.marg_dist()
marginal distribution for x
+---------+----------+
| x_value | x_prob |
+---------+----------+
| 10 | 0.30058 |
| 20 | 0.399609 |
| 30 | 0.299811 |
| sum | 1.0 |
+---------+----------+
marginal distribution for y
+---------+----------+
| y_value | y_prob |
+---------+----------+
| 1 | 0.449896 |
| 2 | 0.550104 |
| sum | 1.0 |
+---------+----------+
d_new.cond_dist()
conditional distribution for x
+---------+---------------------+---------------------+---------------------+-----+
| x_value | 10 | 20 | 30 | sum |
+---------+---------------------+---------------------+---------------------+-----+
| 1 | 0.4453362554901577 | 0.22107998292938813 | 0.33358376158045416 | 1.0 |
| 2 | 0.18219282172098367 | 0.5456168288178236 | 0.2721903494611928 | 1.0 |
+---------+---------------------+---------------------+---------------------+-----+
conditional distribution for y
+---------+---------------------+--------------------+-----+
| y_value | 1 | 2 | sum |
+---------+---------------------+--------------------+-----+
| 10 | 0.6665613147914032 | 0.3334386852085967 | 1.0 |
| 20 | 0.24890080053252053 | 0.7510991994674795 | 1.0 |
| 30 | 0.5005753624783613 | 0.4994246375216386 | 1.0 |
+---------+---------------------+--------------------+-----+
8.13. A Continuous Bivariate Random Vector#
A two-dimensional Gaussian distribution has joint density
We start with a bivariate normal distribution pinned down by
# define the joint probability density function
def func(x, y, μ1=0, μ2=5, σ1=np.sqrt(5), σ2=np.sqrt(1), ρ=.2/np.sqrt(5*1)):
A = (2 * np.pi * σ1 * σ2 * np.sqrt(1 - ρ**2))**(-1)
B = -1 / 2 / (1 - ρ**2)
C1 = (x - μ1)**2 / σ1**2
C2 = 2 * ρ * (x - μ1) * (y - μ2) / σ1 / σ2
C3 = (y - μ2)**2 / σ2**2
return A * np.exp(B * (C1 - C2 + C3))
μ1 = 0
μ2 = 5
σ1 = np.sqrt(5)
σ2 = np.sqrt(1)
ρ = .2 / np.sqrt(5 * 1)
x = np.linspace(-10, 10, 1_000)
y = np.linspace(-10, 10, 1_000)
x_mesh, y_mesh = np.meshgrid(x, y, indexing="ij")
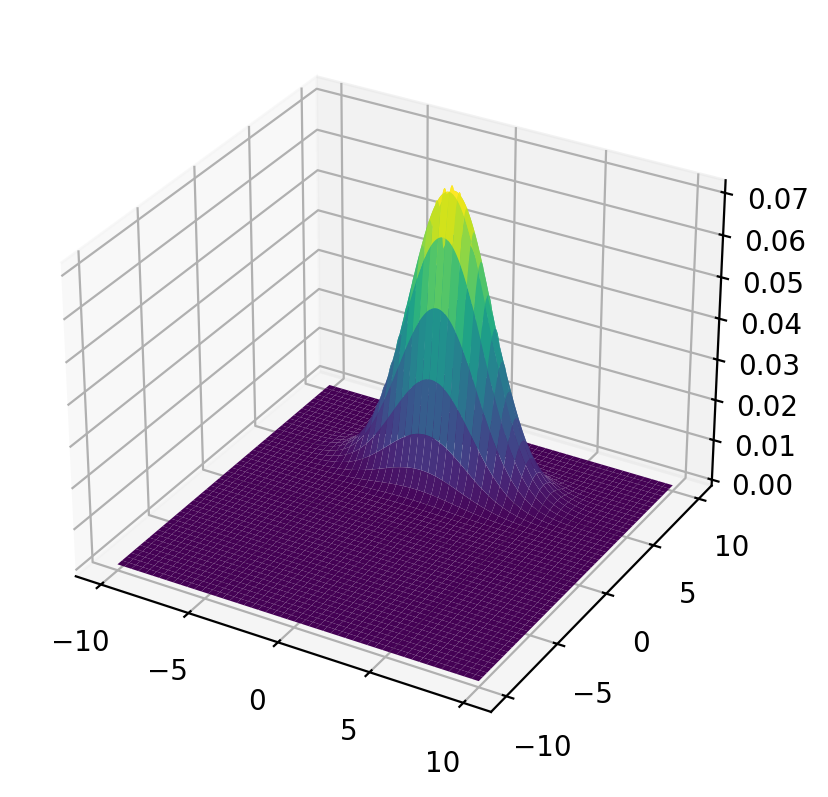
Joint Distribution
Let’s plot the population joint density.
# %matplotlib notebook
fig = plt.figure()
ax = plt.axes(projection='3d')
surf = ax.plot_surface(x_mesh, y_mesh, func(x_mesh, y_mesh), cmap='viridis')
plt.show()
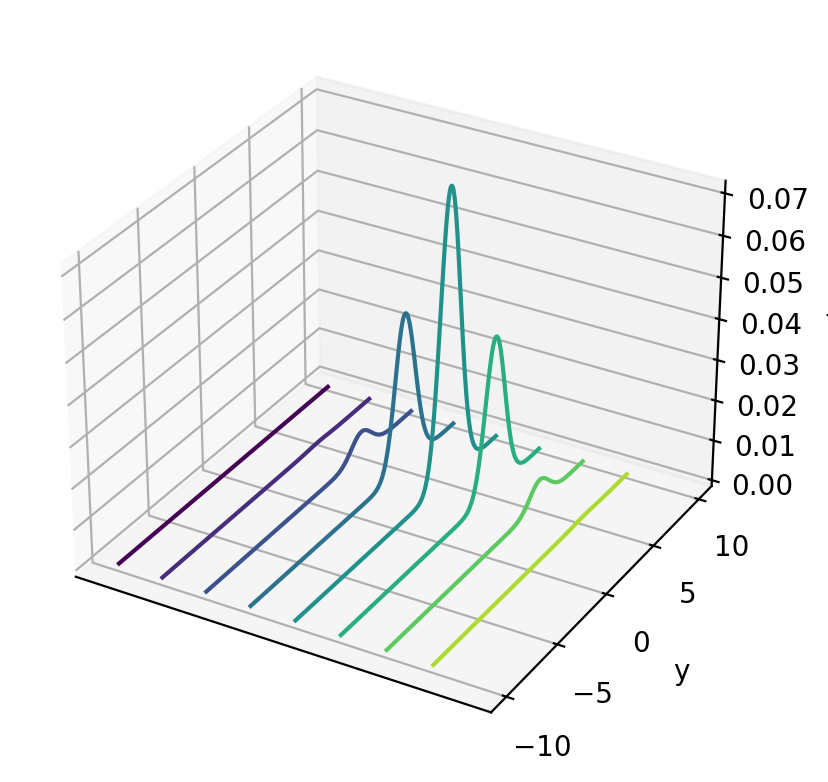
# %matplotlib notebook
fig = plt.figure()
ax = plt.axes(projection='3d')
curve = ax.contour(x_mesh, y_mesh, func(x_mesh, y_mesh), zdir='x')
plt.ylabel('y')
ax.set_zlabel('f')
ax.set_xticks([])
plt.show()
Next we can use a built-in numpy function to draw random samples, then calculate a sample marginal distribution from the sample mean and variance.
μ= np.array([0, 5])
σ= np.array([[5, .2], [.2, 1]])
n = 1_000_000
data = np.random.multivariate_normal(μ, σ, n)
x = data[:, 0]
y = data[:, 1]
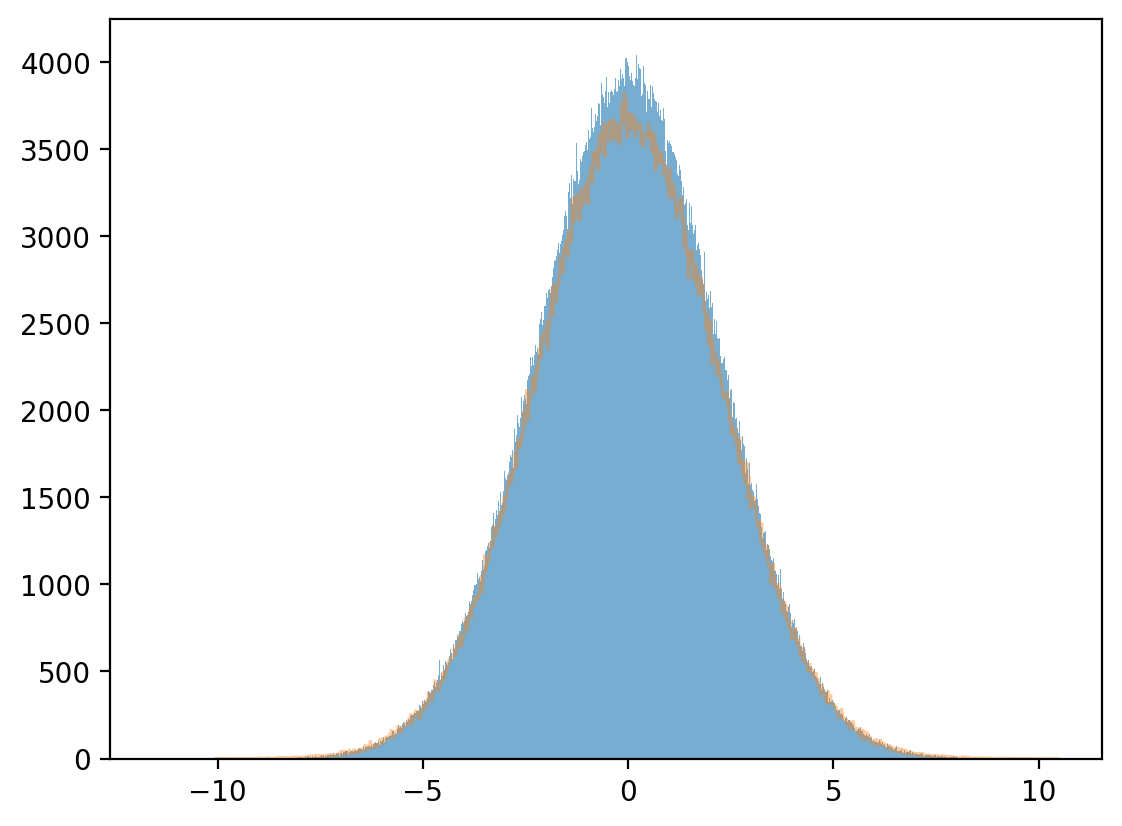
Marginal distribution
plt.hist(x, bins=1_000, alpha=0.6)
μx_hat, σx_hat = np.mean(x), np.std(x)
print(μx_hat, σx_hat)
x_sim = np.random.normal(μx_hat, σx_hat, 1_000_000)
plt.hist(x_sim, bins=1_000, alpha=0.4, histtype="step")
plt.show()
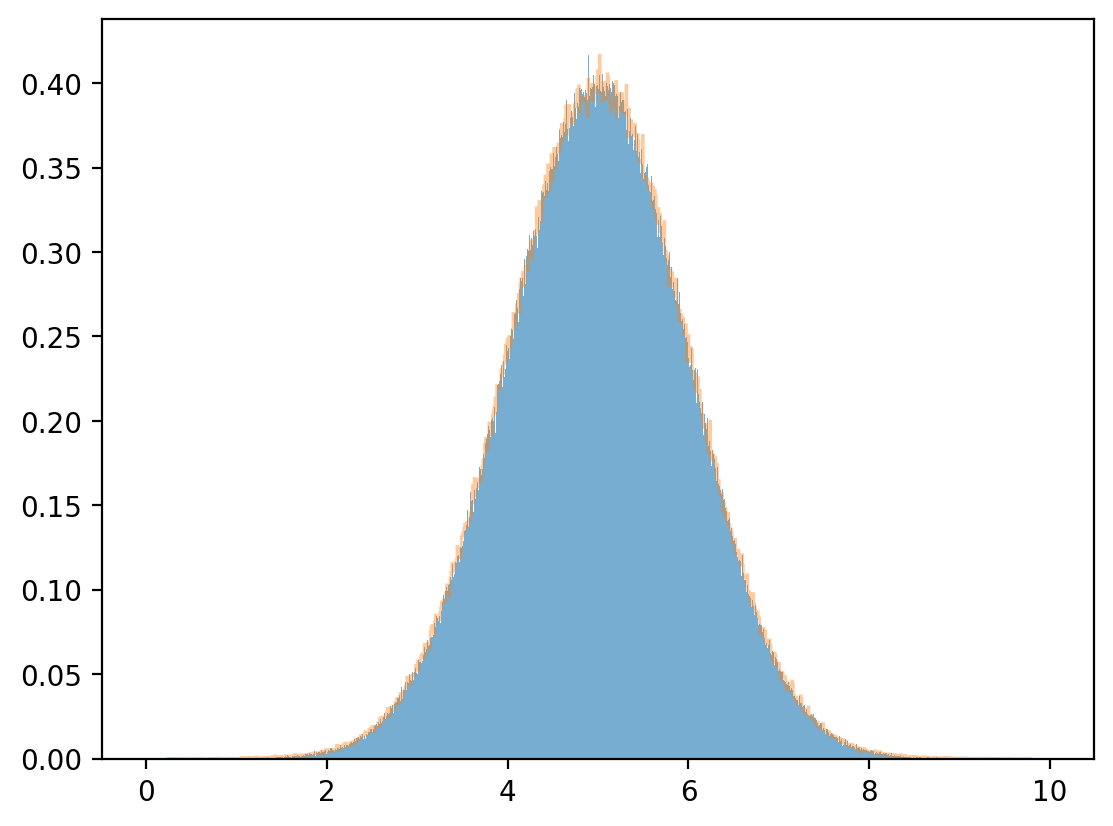
plt.hist(y, bins=1_000, density=True, alpha=0.6)
μy_hat, σy_hat = np.mean(y), np.std(y)
print(μy_hat, σy_hat)
y_sim = np.random.normal(μy_hat, σy_hat, 1_000_000)
plt.hist(y_sim, bins=1_000, density=True, alpha=0.4, histtype="step")
plt.show()
Conditional distribution
For a bivariate normal population distribution, the conditional distributions are also normal:
Note
Please see this quantecon lecture for more details.
Let’s approximate the joint density by discretizing and mapping the approximating joint density into a matrix.
We can compute the discretized marginal density by just using matrix algebra and noting that
Fix
# discretized marginal density
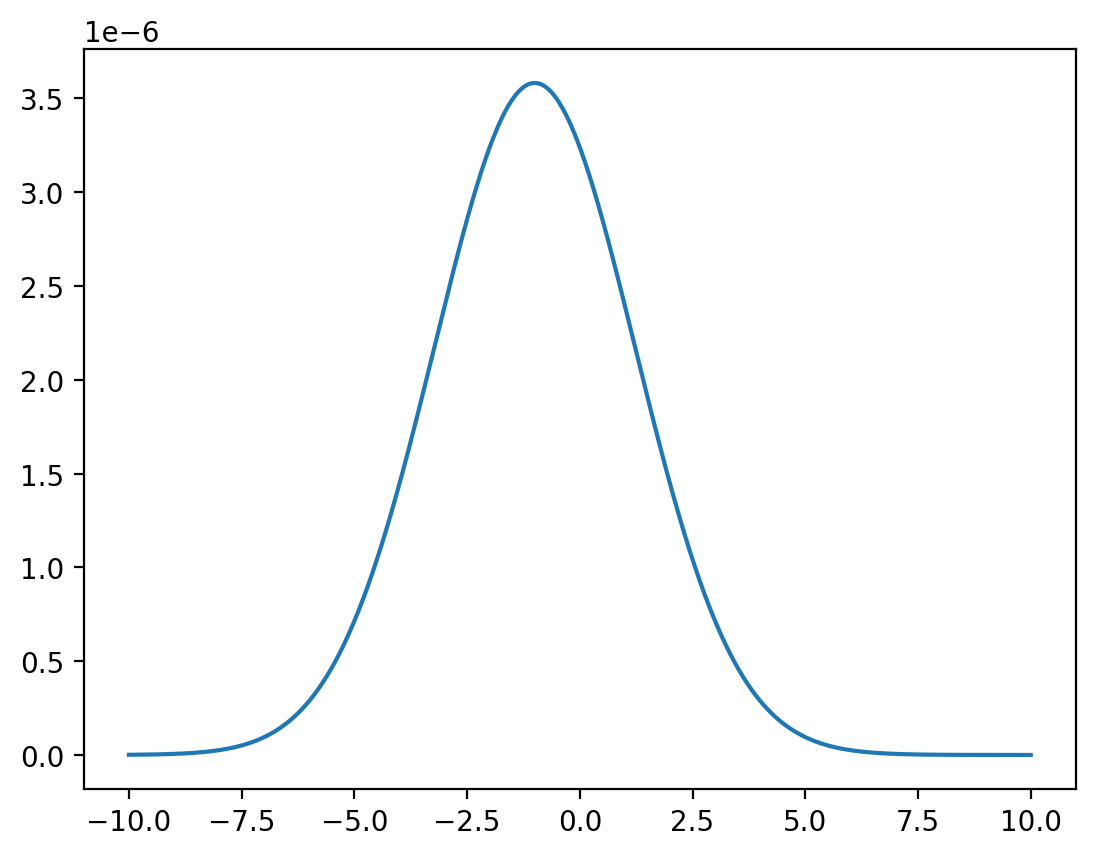
x = np.linspace(-10, 10, 1_000_000)
z = func(x, y=0) / np.sum(func(x, y=0))
plt.plot(x, z)
plt.show()
The mean and variance are computed by
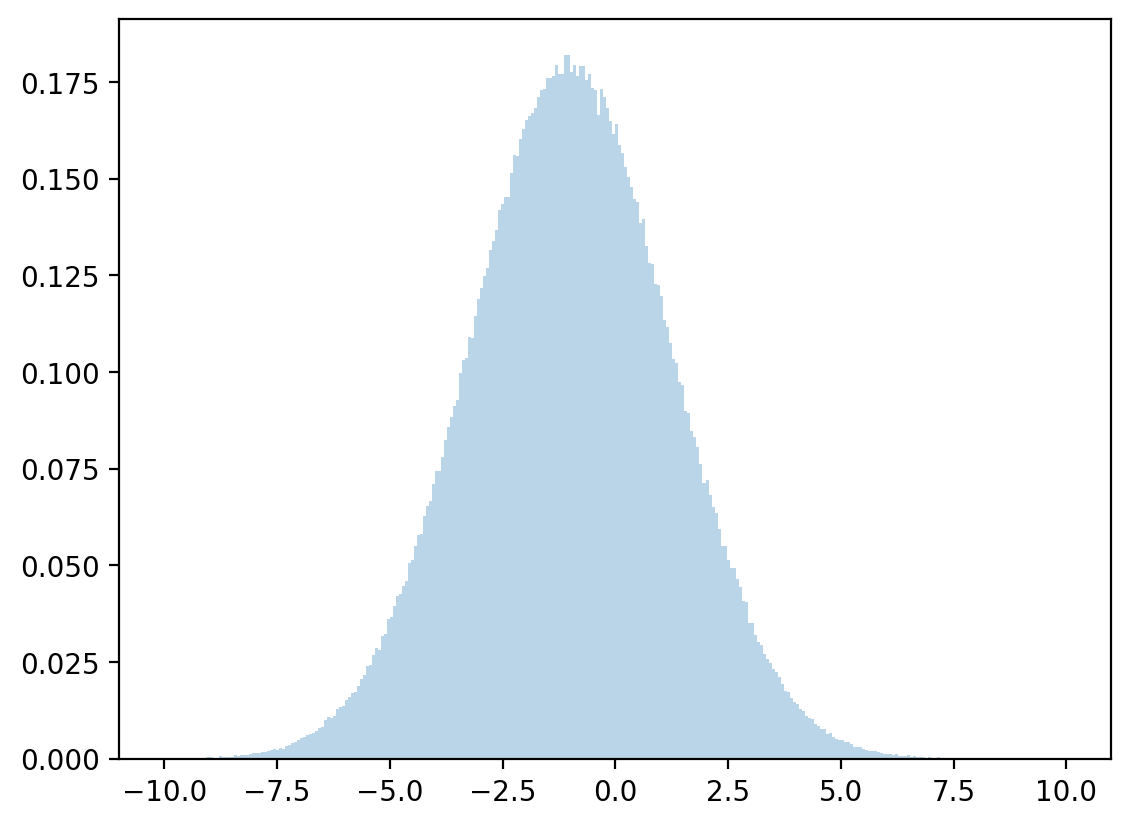
Let’s draw from a normal distribution with above mean and variance and check how accurate our approximation is.
# discretized mean
μx = np.dot(x, z)
# discretized standard deviation
σx = np.sqrt(np.dot((x - μx)**2, z))
# sample
zz = np.random.normal(μx, σx, 1_000_000)
plt.hist(zz, bins=300, density=True, alpha=0.3, range=[-10, 10])
plt.show()
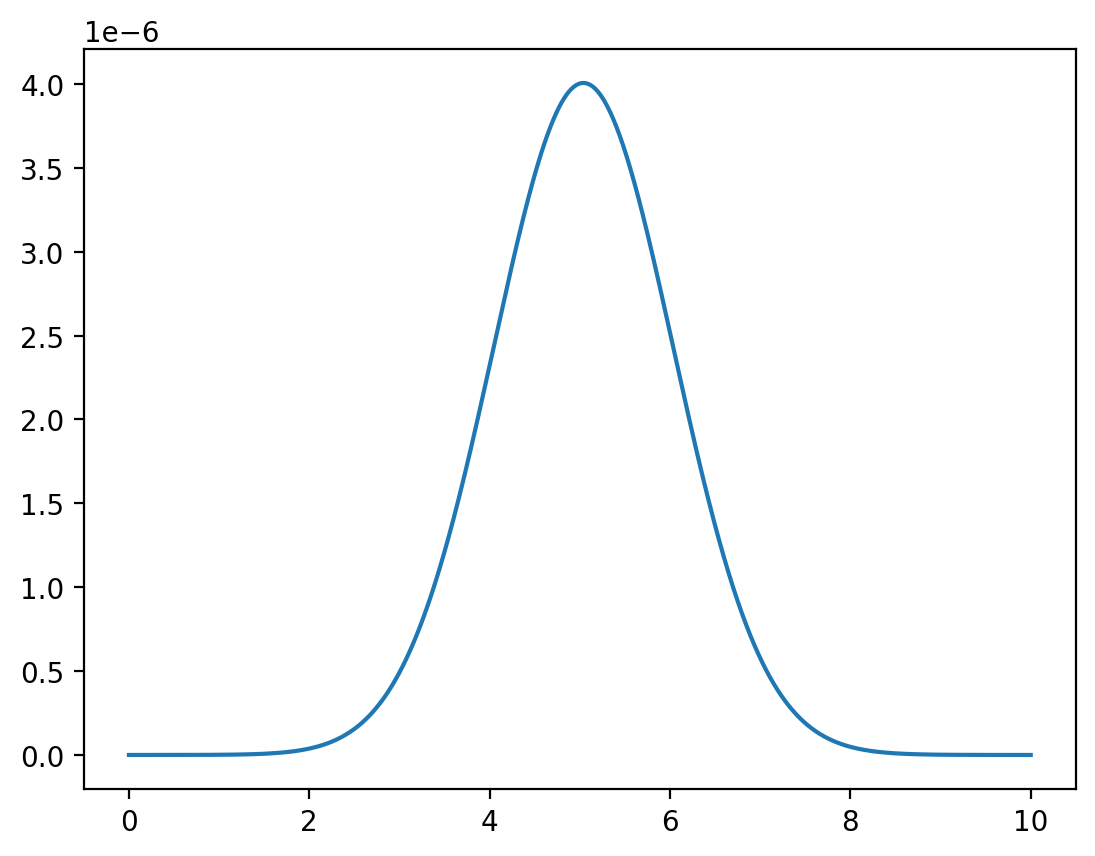
Fix
y = np.linspace(0, 10, 1_000_000)
z = func(x=1, y=y) / np.sum(func(x=1, y=y))
plt.plot(y,z)
plt.show()
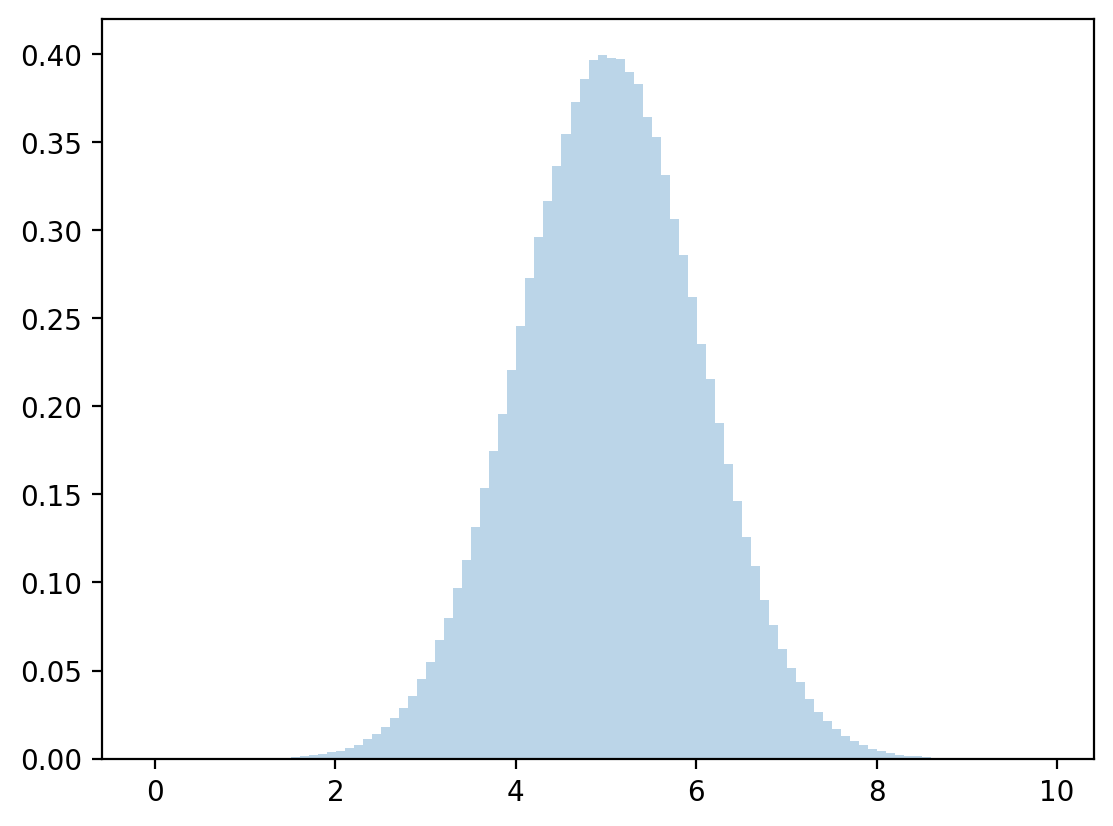
# discretized mean and standard deviation
μy = np.dot(y,z)
σy = np.sqrt(np.dot((y - μy)**2, z))
# sample
zz = np.random.normal(μy,σy,1_000_000)
plt.hist(zz, bins=100, density=True, alpha=0.3)
plt.show()
We compare with the analytically computed parameters and note that they are close.
print(μx, σx)
print(μ1 + ρ * σ1 * (0 - μ2) / σ2, np.sqrt(σ1**2 * (1 - ρ**2)))
print(μy, σy)
print(μ2 + ρ * σ2 * (1 - μ1) / σ1, np.sqrt(σ2**2 * (1 - ρ**2)))
-0.999751841449844 2.2265841331697698
-1.0 2.227105745132009
5.039999456960769 0.9959851265795591
5.04 0.9959919678390986
8.14. Sum of Two Independently Distributed Random Variables#
Let
Define a new random variable
Evidently,
Independence of
Thus, we have:
where
Similarly, for two random variables
where
8.15. Coupling#
Start with a joint distribution
where
From the joint distribution, we have shown above that we obtain unique marginal distributions.
Now we’ll try to go in a reverse direction.
We’ll find that from two marginal distributions, can we usually construct more than one joint distribution that verifies these marginals.
Each of these joint distributions is called a coupling of the two marginal distributions.
Let’s start with marginal distributions
Given two marginal distribution,
Example:
Consider the following bivariate example.
We construct two couplings.
The first coupling if our two marginal distributions is the joint distribution
To verify that it is a coupling, we check that
A second coupling of our two marginal distributions is the joint distribution
The verify that this is a coupling, note that
Thus, our two proposed joint distributions have the same marginal distributions.
But the joint distributions differ.
Thus, multiple joint distributions
Remark:
Couplings are important in optimal transport problems and in Markov processes. Please see this lecture about optimal transport
8.16. Copula Functions#
Suppose that
their marginal distributions are
their joint distribution is
Then there exists a copula function
We can obtain
In a reverse direction of logic, given univariate marginal distributions
Thus, for given marginal distributions, we can use a copula function to determine a joint distribution when the associated univariate random variables are not independent.
Copula functions are often used to characterize dependence of random variables.
Discrete marginal distribution
As mentioned above, for two given marginal distributions there can be more than one coupling.
For example, consider two random variables
For these two random variables there can be more than one coupling.
Let’s first generate X and Y.
# define parameters
mu = np.array([0.6, 0.4])
nu = np.array([0.3, 0.7])
# number of draws
draws = 1_000_000
# generate draws from uniform distribution
p = np.random.rand(draws)
# generate draws of X and Y via uniform distribution
x = np.ones(draws)
y = np.ones(draws)
x[p <= mu[0]] = 0
x[p > mu[0]] = 1
y[p <= nu[0]] = 0
y[p > nu[0]] = 1
# calculate parameters from draws
q_hat = sum(x[x == 1])/draws
r_hat = sum(y[y == 1])/draws
# print output
print("distribution for x")
xmtb = pt.PrettyTable()
xmtb.field_names = ['x_value', 'x_prob']
xmtb.add_row([0, 1-q_hat])
xmtb.add_row([1, q_hat])
print(xmtb)
print("distribution for y")
ymtb = pt.PrettyTable()
ymtb.field_names = ['y_value', 'y_prob']
ymtb.add_row([0, 1-r_hat])
ymtb.add_row([1, r_hat])
print(ymtb)
distribution for x
+---------+----------+
| x_value | x_prob |
+---------+----------+
| 0 | 0.599523 |
| 1 | 0.400477 |
+---------+----------+
distribution for y
+---------+----------+
| y_value | y_prob |
+---------+----------+
| 0 | 0.299864 |
| 1 | 0.700136 |
+---------+----------+
Let’s now take our two marginal distributions, one for
For the first joint distribution:
where
Let’s use Python to construct this joint distribution and then verify that its marginal distributions are what we want.
# define parameters
f1 = np.array([[0.18, 0.42], [0.12, 0.28]])
f1_cum = np.cumsum(f1)
# number of draws
draws1 = 1_000_000
# generate draws from uniform distribution
p = np.random.rand(draws1)
# generate draws of first copuling via uniform distribution
c1 = np.vstack([np.ones(draws1), np.ones(draws1)])
# X=0, Y=0
c1[0, p <= f1_cum[0]] = 0
c1[1, p <= f1_cum[0]] = 0
# X=0, Y=1
c1[0, (p > f1_cum[0])*(p <= f1_cum[1])] = 0
c1[1, (p > f1_cum[0])*(p <= f1_cum[1])] = 1
# X=1, Y=0
c1[0, (p > f1_cum[1])*(p <= f1_cum[2])] = 1
c1[1, (p > f1_cum[1])*(p <= f1_cum[2])] = 0
# X=1, Y=1
c1[0, (p > f1_cum[2])*(p <= f1_cum[3])] = 1
c1[1, (p > f1_cum[2])*(p <= f1_cum[3])] = 1
# calculate parameters from draws
f1_00 = sum((c1[0, :] == 0)*(c1[1, :] == 0))/draws1
f1_01 = sum((c1[0, :] == 0)*(c1[1, :] == 1))/draws1
f1_10 = sum((c1[0, :] == 1)*(c1[1, :] == 0))/draws1
f1_11 = sum((c1[0, :] == 1)*(c1[1, :] == 1))/draws1
# print output of first joint distribution
print("first joint distribution for c1")
c1_mtb = pt.PrettyTable()
c1_mtb.field_names = ['c1_x_value', 'c1_y_value', 'c1_prob']
c1_mtb.add_row([0, 0, f1_00])
c1_mtb.add_row([0, 1, f1_01])
c1_mtb.add_row([1, 0, f1_10])
c1_mtb.add_row([1, 1, f1_11])
print(c1_mtb)
first joint distribution for c1
+------------+------------+----------+
| c1_x_value | c1_y_value | c1_prob |
+------------+------------+----------+
| 0 | 0 | 0.180095 |
| 0 | 1 | 0.41954 |
| 1 | 0 | 0.120156 |
| 1 | 1 | 0.280209 |
+------------+------------+----------+
# calculate parameters from draws
c1_q_hat = sum(c1[0, :] == 1)/draws1
c1_r_hat = sum(c1[1, :] == 1)/draws1
# print output
print("marginal distribution for x")
c1_x_mtb = pt.PrettyTable()
c1_x_mtb.field_names = ['c1_x_value', 'c1_x_prob']
c1_x_mtb.add_row([0, 1-c1_q_hat])
c1_x_mtb.add_row([1, c1_q_hat])
print(c1_x_mtb)
print("marginal distribution for y")
c1_ymtb = pt.PrettyTable()
c1_ymtb.field_names = ['c1_y_value', 'c1_y_prob']
c1_ymtb.add_row([0, 1-c1_r_hat])
c1_ymtb.add_row([1, c1_r_hat])
print(c1_ymtb)
marginal distribution for x
+------------+--------------------+
| c1_x_value | c1_x_prob |
+------------+--------------------+
| 0 | 0.5996349999999999 |
| 1 | 0.400365 |
+------------+--------------------+
marginal distribution for y
+------------+---------------------+
| c1_y_value | c1_y_prob |
+------------+---------------------+
| 0 | 0.30025100000000005 |
| 1 | 0.699749 |
+------------+---------------------+
Now, let’s construct another joint distribution that is also a coupling of
# define parameters
f2 = np.array([[0.3, 0.3], [0, 0.4]])
f2_cum = np.cumsum(f2)
# number of draws
draws2 = 1_000_000
# generate draws from uniform distribution
p = np.random.rand(draws2)
# generate draws of first coupling via uniform distribution
c2 = np.vstack([np.ones(draws2), np.ones(draws2)])
# X=0, Y=0
c2[0, p <= f2_cum[0]] = 0
c2[1, p <= f2_cum[0]] = 0
# X=0, Y=1
c2[0, (p > f2_cum[0])*(p <= f2_cum[1])] = 0
c2[1, (p > f2_cum[0])*(p <= f2_cum[1])] = 1
# X=1, Y=0
c2[0, (p > f2_cum[1])*(p <= f2_cum[2])] = 1
c2[1, (p > f2_cum[1])*(p <= f2_cum[2])] = 0
# X=1, Y=1
c2[0, (p > f2_cum[2])*(p <= f2_cum[3])] = 1
c2[1, (p > f2_cum[2])*(p <= f2_cum[3])] = 1
# calculate parameters from draws
f2_00 = sum((c2[0, :] == 0)*(c2[1, :] == 0))/draws2
f2_01 = sum((c2[0, :] == 0)*(c2[1, :] == 1))/draws2
f2_10 = sum((c2[0, :] == 1)*(c2[1, :] == 0))/draws2
f2_11 = sum((c2[0, :] == 1)*(c2[1, :] == 1))/draws2
# print output of second joint distribution
print("first joint distribution for c2")
c2_mtb = pt.PrettyTable()
c2_mtb.field_names = ['c2_x_value', 'c2_y_value', 'c2_prob']
c2_mtb.add_row([0, 0, f2_00])
c2_mtb.add_row([0, 1, f2_01])
c2_mtb.add_row([1, 0, f2_10])
c2_mtb.add_row([1, 1, f2_11])
print(c2_mtb)
first joint distribution for c2
+------------+------------+----------+
| c2_x_value | c2_y_value | c2_prob |
+------------+------------+----------+
| 0 | 0 | 0.299782 |
| 0 | 1 | 0.299538 |
| 1 | 0 | 0.0 |
| 1 | 1 | 0.40068 |
+------------+------------+----------+
# calculate parameters from draws
c2_q_hat = sum(c2[0, :] == 1)/draws2
c2_r_hat = sum(c2[1, :] == 1)/draws2
# print output
print("marginal distribution for x")
c2_x_mtb = pt.PrettyTable()
c2_x_mtb.field_names = ['c2_x_value', 'c2_x_prob']
c2_x_mtb.add_row([0, 1-c2_q_hat])
c2_x_mtb.add_row([1, c2_q_hat])
print(c2_x_mtb)
print("marginal distribution for y")
c2_ymtb = pt.PrettyTable()
c2_ymtb.field_names = ['c2_y_value', 'c2_y_prob']
c2_ymtb.add_row([0, 1-c2_r_hat])
c2_ymtb.add_row([1, c2_r_hat])
print(c2_ymtb)
marginal distribution for x
+------------+--------------------+
| c2_x_value | c2_x_prob |
+------------+--------------------+
| 0 | 0.5993200000000001 |
| 1 | 0.40068 |
+------------+--------------------+
marginal distribution for y
+------------+-----------+
| c2_y_value | c2_y_prob |
+------------+-----------+
| 0 | 0.299782 |
| 1 | 0.700218 |
+------------+-----------+
We have verified that both joint distributions,
So they are both couplings of